Dr. Azra Frkatović-Hodžić from Genos Glycoscience Research Laboratory in Zagreb, Croatia, discusses a research paper she co-authored that was published by Aging (Aging-US) in Volume 15, Issue 24, entitled, “Mapping of the gene network that regulates glycan clock of ageing.”
Behind the Study is a series of transcribed videos from researchers elaborating on their recent studies published by Aging (Aging-US). Visit the Aging (Aging-US) YouTube channel for more insights from outstanding authors.
—
Hello, my name is Azra Frkatovic-Hodzic, and I’m a researcher in Genos. Genos is a company which specializes in the field of glycomics research. In our recently published paper in Aging journal titled as, “Mapping of the gene network that regulates glycan clock of ageing,” we present our work done on exploring the genetic regulation of IgG galactosylation.
So what is galactosylation or glycosylation in general? Glycosylation is post-translational modification that is characterized by the attachment of oligosaccharide chains to a protein or a lipid. Now these oligosaccharide chains we usually call as glycans. Glycans that are linked to protein and specifically to IgG, actually largely affect function of IgG in the immune response. If the glycan is remote from IgG, actually IgG becomes or completely loses its activity in the immune system.
Now, if we talk about presence of galactose units in IgG glycans, dozens of studies have shown that actually absence of galactose is associated with inflammation in humans, this inflammation either stemming from the disease, but also with advanced aging in humans.
This is why it’s important that we understand in what way this process is regulated and several studies have now shown that IgG glycosylation is largely influenced by genetics. So up to 75% of our variation that we observe in IgG glycans in humans can be explained by a genetic component. So given that we have access to IgG population data, IgG glycan population data, we were able to run genome wide association studies as means of exploring the genetic background of variation in IgG glycosylation.
In this study, we focus specifically on galactosylation phenotypes. So either absence of galactose, presence of one galactose and presence of two galactoses, so monogalactosylation and the galactosylation.
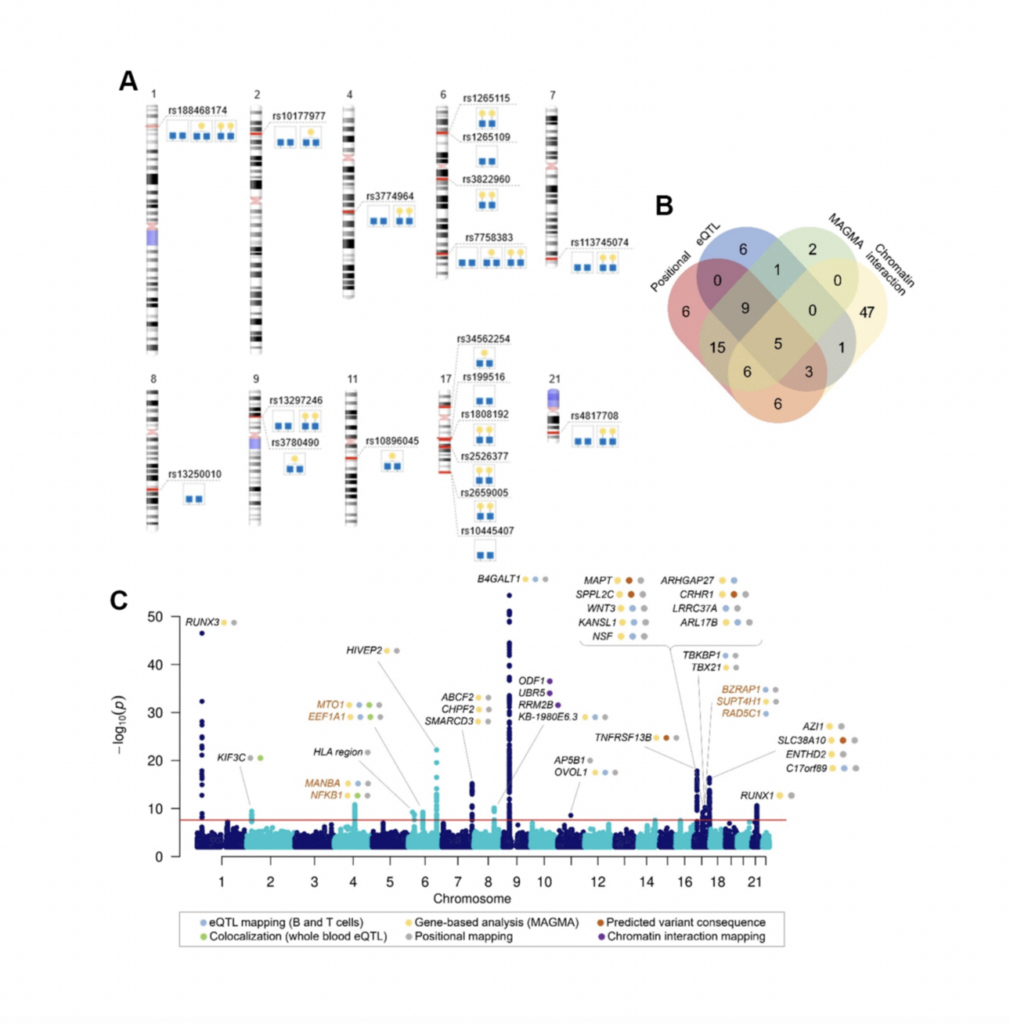
In this study, we had around 13,700 samples in total after meta-analysis, and we identified 16 genomic regions out of which three were considered novel. Then we proceeded by prioritizing genes in those 16 genomic regions in order to get the list of genes to be experimentally validated. We narrowed down this list to seven genes, and we assessed their functional role in IgG galactosylation in HEK293 free style expression system based on the CRISPR-cas9 molecular tools. The system also was transfected with plasma that contains IgG light and heavy chain genes in order to also excrete IgG as the system does not inherently excrete IgG.
So we selected seven genes and then did both up regulation and down regulation of their expression. The results of the study now showed that MANBA, TNFRSF13B and EEF1A1 gene expression resulted in the changes in of IgG galactosylation levels, specifically up regulation of MANBA gene expression resulted in decrease of IgG monogalactosylated species.
Now why is this interesting? It’s interesting because this MANBA gene encodes a beta-mannosidase, which is an enzyme that cleaves beta-monos units from the non-reducing end of IgG and glycans in general, but it was not previously associated with IgG glycans specifically. So it has a known role in glycosylation process, but also what is interesting is that mutations in MANBA are known to affect kidney function, blood pressure, and also cardiovascular disease risk. On the other side, we have monogalactosylated IgG glycans that are also associated and identified as the best predictors of future cardiovascular events in women.
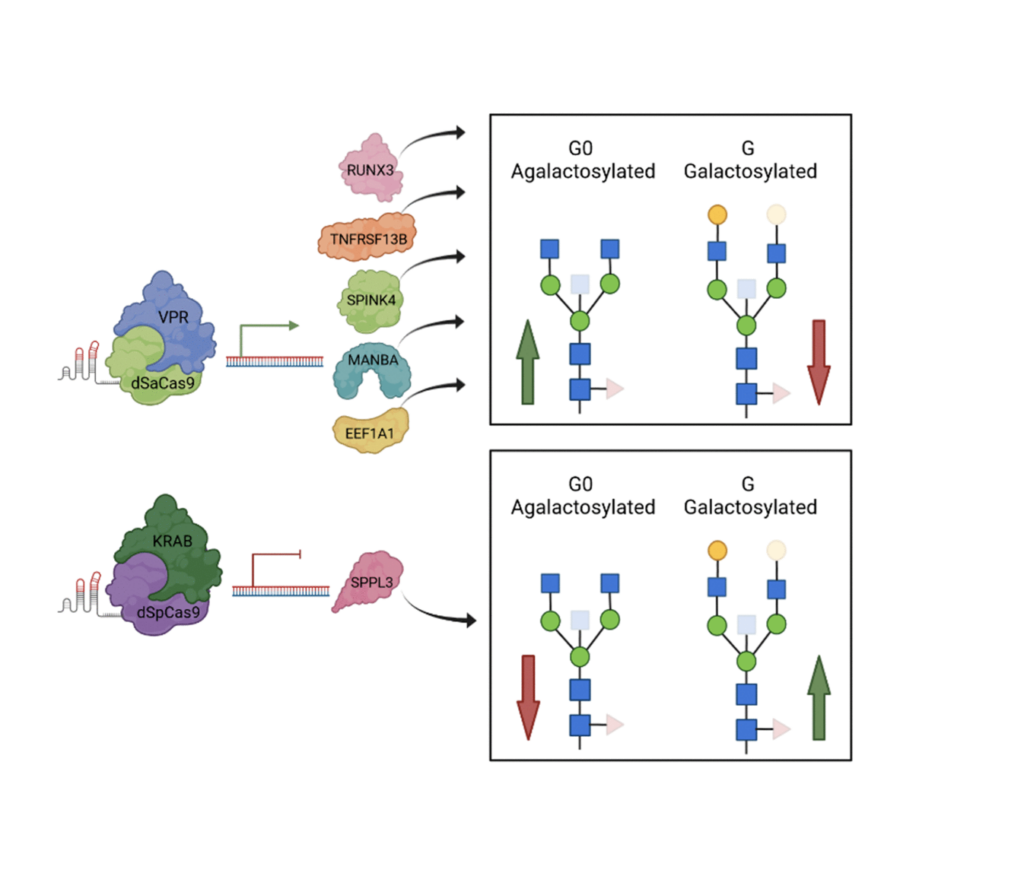
Now, since MANBA expression was affecting the monogalactosylated IgG glycan levels, we could hypothesize that there is a link between MANBA galactose and cardiovascular events. However, at this point, we do not know these mechanisms that could explain this link, but also this really warrants our future studies. Also, we had the increase of the EEF1A1 expression levels were increased. We could observe decrease in galactosylated IgG glycans. Now interestingly, EEF1A1, was associated with a pro-inflammatory modulation of IgG, but now we can hypothesize that it could happen by decreasing its galactosylation.
The role of other genes, unfortunately, could not be validated in the HEK293 freestyle, the transit expression system, because it has also some of the limitations. We have to remember that this system does not inherently secrete immunoglobulins and epigenetic context of these cells is not equivalent to that of plasma and B cells, where glycosylation process actually occurs.
In addition, it could be that the alternative galactosylation might depend on the gene expression in other cell types, and also interactions between cells such as B and T cells, so we might not always observe the natural effects of the gene expression manipulation, because we don’t have the right biological context. However, we do plan to expand our GWAS studies by increasing the sample sizes, but also continuing to work on developing the efficient system for functional validation of our GWAS hits, both for galactosylation but also other features of IgG glycans.
And at the end, I want to thank my mentors Gordon Lauc, Lucija Klaric, and Frano Vucković for their support, my co-authors, our collaborators and colleagues. And I hope we’ll continue to work on exploring this important process together in future. Thank you.
Click here to read the full study published by Aging (Aging-US).
—
Aging (Aging-US) is an open-access journal that publishes research papers bi-monthly in all fields of aging research and other topics. These papers are available to read at no cost to readers on Aging-us.com. Open-access journals offer information that has the potential to benefit our societies from the inside out and may be shared with friends, neighbors, colleagues, and other researchers, far and wide.
For media inquiries, please contact [email protected].